Molybdenum cofactor deficiency: Molecular basis, diagnosis and therapy
Assimilatory NAD(P)H:nitrate reductases (NR, EC 1.7.1.1-3) catalyze the first and rate-limiting step of nitrate assimilation in plants, algae and fungi. NR belongs to the molybdenum cofactor- (Mo-cofactor) containing family of enzymes that catalyze two-electron transfer reactions in global metabolic cycles including nitrate reduction, phytohormone synthesis, purine catabolism, sulfite detoxification and many others.
Reversible inactivation of plant NR is initiated by phosphorylation of a conserved serine located between the molybdenum domain, where catalysis takes place, and the heme domain followed by binding of a 14-3-3 protein. Our goal is to identify the functional and structural mechanism by which NR activity is turned down via 14-3-3 proteins by using a fully defined in vitro system. We will characterize the biophysical, enzymatic and structural properties of the apo-, the phosphorylated as well as 14-3-3-complexed enzyme.
Projects
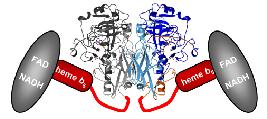
Structure of Arabidopsis nitrate reductase
We have determined the crystal structures of plant sulfite oxidase and yeast nitrate reductase catalytic domain. Here we wish to complete our structural knowledge about plant nitrate reductase by solving the holo-structure. These studies should uncover the path of intra-molecular electron transfer and will help to understand the entire catalytic cycle of nitrate reduction.
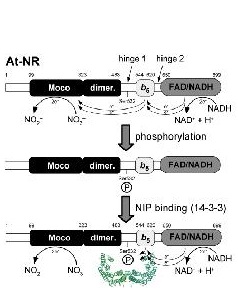
Functional and structural basis of 14-3-3 mediated inhibition of nitrate reductase
Reversible inactivation of plant NR is initiated by phosphorylation of a conserved serine located between the molybdenum domain, where catalysis takes place, and the heme domain followed by binding of a 14-3-3 protein. Our goal is to identify the functional and structural mechanism by which NR activity is turned down via 14-3-3 proteins by using a fully defined in vitro system. We have already cloned, expressed and purified 11 different plant 14-3-3 isoforms and could show that only some of them bind and inhibit NR with high affinity. Question to be answered are:
- Which step in the catalytic cycle is inhibited by NR phosphorylation and 14-3-3 protein binding?
- What are the binding affinities of different Arabidopsis 14-3-3 proteins to phosphorylated NR?
- What is the structural basis of 14-3-3-mediated NR inhibition?
Funding
- DFG SFB 635 (TP05)
Tools
- standard tools in molecular biology and biochemistry (PCR, molecular cloning, Western blots, enzyme assays, etc.)
- recombinant protein expression in E. coli, yeast and yeast (fermentation)
- protein purification by affinity chromatography (IMAC, GST, intein, strep, etc.) and standard chromatography
- steady state enzyme kinetics (photometric, ELISA) and UV/vis spectroscopy
- protein interaction studies (isothermal titration calorimentry, complex formation and purification)
- phosphorylation assays and isolation of phospho-proteins
- protein crystallography and structural biology
Collaboration
- Prof. Dr. Russ Hille
Department of Biochemistry and Cell Biology, University of California at Riverside, USA - Dr. Markus Teige
Max F. Perutz Laboratories, Institute for Biochemistry, University Vienna, Austria - Prof. Dr. Wilbur Campbell
Michigan Tech, USA
Project-relevant publications
- Fischer, K., Barbier, GG, Hecht, HJ, Mendel, RR, Campbell, WH, Schwarz,G.* 2005
Crystal Structure of the Yeast Nitrate Reductase Molybdenum Domain Provides Insight into Eukaryotic Nitrate Assimilation.
Plant Cell 17, 1167-1179 - Schwarz, G., and Mendel, R. R. 2006.
Molybdenum Cofactor Biosynthesis and Molybdenum Enzymes.
Annu Rev Plant Biol. 57: 623-647 - Schrader, N., Fischer, K., Theis, K., Mendel, R.R., Schwarz, G.*, and Kisker, C. 2003.
The Crystal Structure of Plant Sulfite Oxidase Provides Insights into Sulfite Oxidation in Plants and Animals.
Structure, 11, 1251-1263 - Eilers, T., Schwarz, G., Brinkmann, H., Witt, C., Richter, T., Nieder, J., Koch, B., Hille, R., Hansch, R., and Mendel, R.R. 2001.
Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism.
J. Biol. Chem. 276:46989-46994.